Two faculty members, Dr. David Rasmussen and Dr. Benjamin Callahan, were awarded USDA NIFA grants through a new program called FACT: Food and Agricultural Cyberinformatics and Tools Initiative.
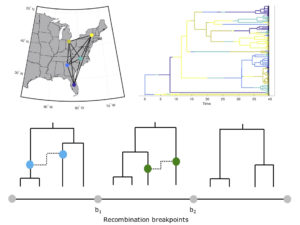
Dr. Rasmussen’s project is called, “Next Generation Spatial Epidemiology for Tracking the Spread of Plant Pathogens”. The main goal is: “To develop the next generation of phylogenetic tools for tracking the spread of plant pathogens ranging from viruses to fungi through complex agricultural landscapes. Since many of these pathogens undergo occasional recombination, another goal is to use information about recombination events across pathogen genomes to identify the spatial location of historical recombination events and the geographic source of particular genes involved in pathogenesis.”
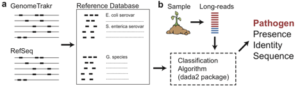
Dr. Callahan’s project is called, “Rapid Detection and Tracking of Foodborne Pathogens with Long-read Amplicon Sequencing”. Food-borne pathogens enact substantial harms on the American people in the form of illness, lost productivity, and expenses related to mitigation and regulatory compliance. Surveillance and tracing of foodborne pathogens is a key control strategy, but its efficacy is reduced by the long-times associated with current culture and whole-genome-sequencing approaches. Rapid, accurate and comprehensive pathogen detection would improve the safety and lower the costs of our food supply.
We aim to develop a targeted metagenomics methodology that can rapidly (<24 hrs) and precisely identify a broad range of foodborne pathogens from heterogeneous environmental samples. In order to achieve this, we propose to combine the GenomeTrakr and NCBI RefSeq databases with cutting-edge bioinformatics tools developed by the PD that achieve single-nucleotide resolution from amplicon sequencing data of full-length genes to identify E. coli and Salmonella strains to the serovar level (e.g. E. coli O157:H7 or S. enterica Heidelberg). We will validate the resolution and accuracy of this new methodology in silico, on isolates of various pathogenic serovars, and in environmental samples of various types for which pathogen presence and identity were previously established by standard culture-based methods. Our methodology will be distributed to the broader food safety community as open-source and actively-supported software, alongside extensive documentation of its efficacy and best-practices guidance. Successful completion of this project will yield a powerful, usable, and broad-spectrum pathogen surveillance technique that will improve food safety by detecting foodborne pathogens before they reach consumers, and by rapidly tracing outbreaks to their source.”