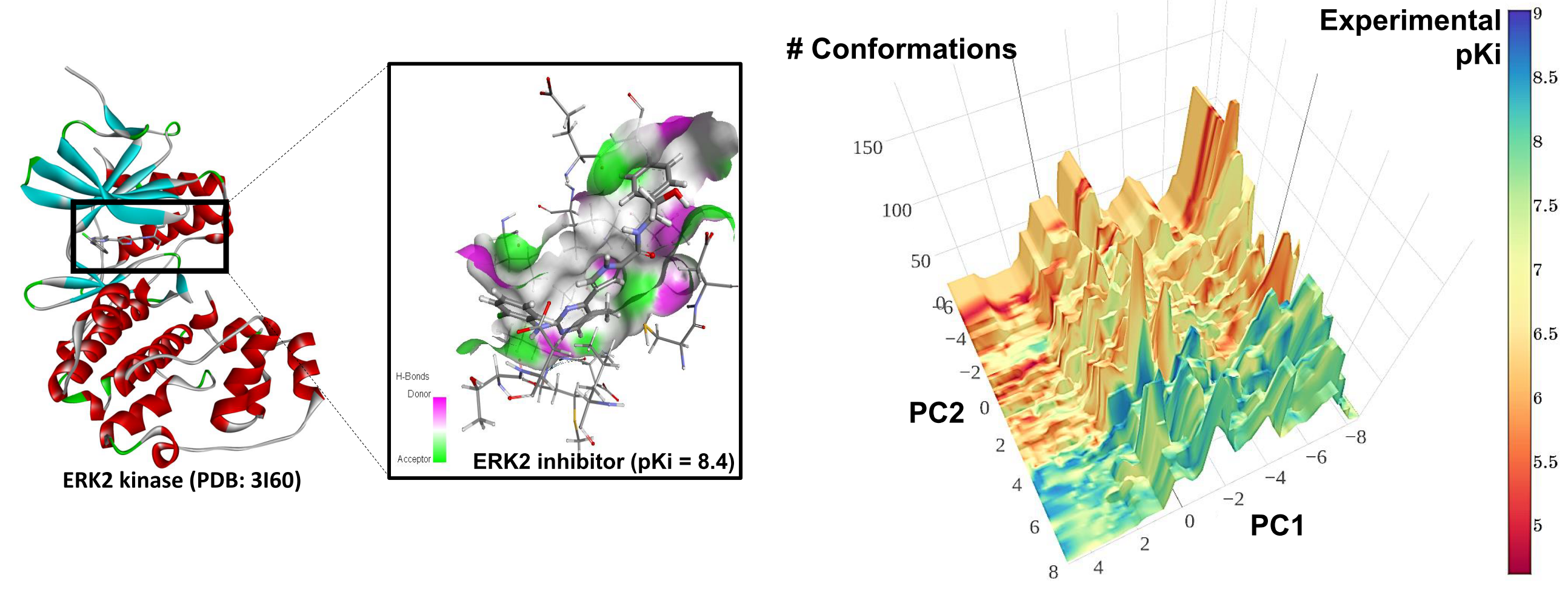
It’s 2015, and the link between inherited DNA variation and numerous diseases is well-established. However, an important question remains – how, exactly do these links between genetic variation and disease work? The Genotype-Tissue Expression (GTEx) project, funded by the National Institutes of Health and including a team of researchers from NC State and UNC-Chapel Hill, aims to start answering that question by looking at how genetic variation affects gene expression.
“Important gaps still remain in understanding genetic processes, which vary greatly across the organs and tissues of the human body,” says Fred Wright, professor of statistics and biological sciences at NC State. “We have little understanding of how genetic variants actually cause disease, because we haven’t been able to look at the gene expression part of the equation. GTEx aims to fill the knowledge gap between the DNA you’re born with and actual disease outcomes.”
“You can think of DNA as the controller of a giant genetic switchboard,” explains Wright’s collaborator Andrew Nobel, professor of statistics and operations research at UNC-Chapel Hill. “When DNA switches on a gene, the gene produces proteins with specific functions. In the case of many common diseases, relatively small changes in protein output can have profound effects on disease risk.”
The GTEx project took samples of a large variety of tissues from 175 recently deceased individuals, measuring gene expression in those tissues. First, the researchers established that nearly normal gene activity persists for several hours after death. Then the major task of connecting variation in DNA to expression began. This is where Wright, Nobel and their team came in – to find meaningful correlations among all the “noise.”
“We had data for millions of DNA variants and how each variant was related to gene expression in different tissues,” Wright says. “Since we were looking at multiple tissues, there were gaps and overlaps in the data. We had to come up with a mathematical/statistical model that could assess, for each DNA variant and each gene, the evidence for the variant-gene combination being active for each of the tissues. The data were analyzed all together, but we untangled associations that reflected underlying true biology, versus associations that were happenstance because of sample overlap.”
Initial results were promising. The group found that DNA variants that affect expression tend to do so either in one tissue alone, or in all the examined tissues. Groups within GTEx are now comparing the results of the model to each variant-disease association, helping us further narrow down the genes that the variants affect, and in which tissue. The work may bring us one step closer to personalized therapies for numerous diseases.
The results appear in Science (DOI: 10.1126/science.1262110). NC State co-authors include Yi-Hui Zhou, research assistant professor of biological sciences. Funding was provided by the National Institutes of Mental Health and the NIH Common Fund.