Researchers from North Carolina State University have released the first chemical “map” of dyes from the Max A. Weaver Dye Library, which contains almost 100,000 samples of unique dyes and fabrics. The information could assist researchers in developing dyes with desirable properties.
NC State analytic chemist Nelson Vinueza is working on digitizing and analyzing the library so that its contents are accessible to the public. “Each vial has the chemical structure written on it, so we must first digitize those molecular structures and then select candidates to do further characterization,” Vinueza says. “Obviously with a library of this size, the time and expense associated with characterizing each dye would be prohibitive, so we needed a faster, more efficient way to be able to analyze these dyes.”
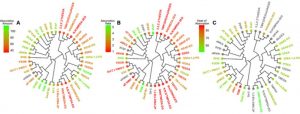
Vinueza partnered with NC State computational chemist Denis Fourches to create a cheminformatics map of the 2,700 dyes that had their molecular structures already digitized. The computer models allowed the researchers to compare dyes with similar chemical structures and properties.
The cheminformatics analysis also enabled the identification of 150 chemically unique dyes representative of the library. In order to assist researchers in developing dyes with desirable properties, these sampled chemical structures are now publicly available in the ChemSpider database (www.chemspider.com/DatasourceDetails.aspx?id=900). “There are 58 million chemicals in the ChemSpider database, and 143 of the dyes have completely unique chemistry, which is really fantastic,” Vinueza says.
“We believe that this addition can prove invaluable to researchers who are looking for particular characteristics in these chemicals, such as antibiotic or anti-cancer properties, or for dyes that absorb light in ways that could lead to better solar-cell technology,” Vinueza continues. “This dye library could prove invaluable in creating cutting-edge solutions to problems ranging from human health to the environment.”
“The chemical maps and the other cheminformatics modeling techniques we used here provide a cheaper and faster way to screen for chemical dyes with the desired properties,” Fourches says. “Doing that same analysis by experimentally characterizing and testing all samples in a lab would take decades.
“And since these dyes were constructed sequentially over time, it is straightforward to pinpoint where structural changes led to properties of interest. We actually show that small modifications of the dyes’ chemical structures can lead to dramatic changes of their properties. This library is a real treasure trove for chemists.”
The research appears in Chemical Science, and was funded by NC State Chancellor’s Faculty Excellence program. The Max A. Weaver Dye Library was donated to the NC State College of Textiles in 2014 by the Eastman Chemical Company. Vinueza and Fourches are co-corresponding authors. Postdoctoral scholar Melaine Kuenemann in the Fourches laboratory is lead author. NC State graduate students Yufei Chen and Nadia Sultana from Vinueza’s laboratory, research assistant professor Malgorzata Szymczyk, Ciba professor of Dye Chemistry Harold Freeman, Dean David Hinks, and the Environmental Protection Agency’s Antony Williams contributed to the work.
“Weaver’s Historic Accessible Collection of Synthetic Dyes: A Cheminformatics Analysis”
DOI: 10.1039/C7SC00567A