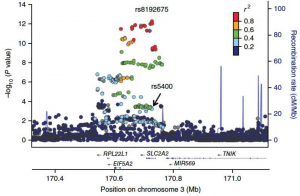
Drs. Daniel Rotroff and Alison Motsinger-Reif, in collaboration with the international Metformin Genetics (MetGen) Consortium, have uncovered new genetic evidence of how the benefits of the world’s most commonly used Type 2 diabetes drug, Metformin, may vary between individuals. Metformin is the first-line antidiabetic drug and has over 100 million users worldwide, yet its mechanism of action remains unclear. The study conducted a genome-wide association study (GWAS) that identified a genetic variant in the gene encoding the glucose transporter GLUT2, a protein that plays an important role in transporting glucose inside the body. They showed that those people who carried this variant had reduced levels of GLUT2 in the liver and other tissues resulting in a defect in how the body handles glucose. Metformin acted to specifically reverse this deficiency resulting in a better response to metformin in people carrying this gene variant. This is the largest precision medicine study on an anti-diabetic drug performed to date, and represents an exciting step towards personalized therapy for Type 2 diabetes.
The entire publication can be viewed here: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5007158/